MCB TRANSCRIPT
Gene Expression: What Viruses Can Tell Us About Ourselves

Viruses use us for their own devices, co-opting our cellular machinery to replicate and spread. Likewise, MCB Associate Professor Britt Glaunsinger bests viruses at their own game by making them work for us. "They're like a flashlight that can illuminate how gene expression works," she says. "They're not inventors, they're thieves and they've evolved to steal the really important things."
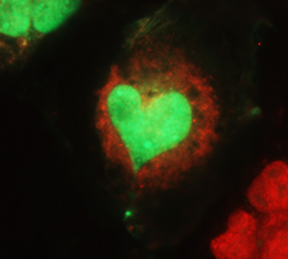
protein (red) distribution between the cytoplasm
and the nucleus in monkey cells.
The virus she uses is Kaposi's sarcoma-associated herpesvirus (KSHV), which limits gene expression by attacking mRNAs. "It makes an enzyme that slices mRNAs in half — they're dead, they can't make proteins," says Glaunsinger, who made this discovery as a post-doc.
Now, she's mapping binding between viral and cellular proteins. These interactions should reveal how cellular pathways like those for gene regulation work, as well as how viruses hijack cell components to, for example, escape our immune systems.
Intriguingly, cells may also co-opt viral strategies. Glaunsinger has found that KSHV infection swamps cells with short RNAs from non-coding DNA sequences, and she suspects cells then use those short RNAs to activate pathways that signal the virus's presence.
"Our goal is to discover how we work and how they work," Glaunsinger says. "Discovering a virus's Achilles heel would benefit human health."
Protein Regulation: How Cells Break Down Proteins

When proteins are damaged or unneeded, cells destroy them in a molecular machine — the proteasome — that recycles the amino acids. The proteasome regulates protein levels and "protein levels control cell functions," says MCB Associate Professor Andreas Martin.
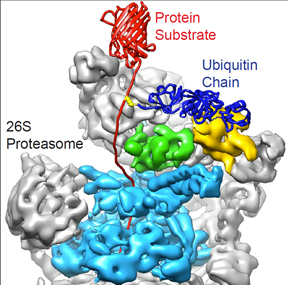
with ubiquitin chains (dark blue) and targeted
for destruction by the 26S proteasome, which
has a ring-shaped molecular motor (cyan) that
unravels targeted proteins and stuffs them into
an internal chamber where they are broken down.
The proteasome has a molecular motor that unravels proteins flagged for destruction, and then pulls them into a chamber with catalytic active sites (proteases) that break them down. The proteasome is made of more than 35 protein subunits, and by using in vitro biochemistry and cryo-electron microscopy Martin was the first to discover exactly how they fit together.
"Now we can introduce specific mutations that disrupt particular features," Martin says, explaining that this will reveal what various parts of the proteasome do and how they are regulated. His lab uses structural, biochemical and biophysical studies to assess how proteins are degraded, and single molecule experiments to investigate mechanical protein unraveling. "We can measure the forces applied by the motor, and the length increment of the protein that gets tugged into the chamber during every power stroke," Martin says.
Proteasomes control cell division, making them a target for cancer treatments, and proteasome inhibitors are approved for treating multiple myeloma and a type of lymphoma. Ultimately, Martin's work could help researchers fine-tune these cancer therapies.




